Product Cat. No.: GBS-002
For Research Use Only.
Human ALK gene fusion detection kit.
10 Tests/box.
This kit uses fluorescence in situ hybridization to detect the fusion status of ALK gene in non-small cell lung cancer. The detection samples are surgical resection or biopsy samples of non-small cell lung cancer embedded in 4% neutral formaldehyde fixed paraffin. The product has not been clinically verified in combination with ALK targeted therapeutic drugs, and its clinical detection performance has been confirmed by comparative test with accompanying diagnostic reagents that have been verified by targeted drugs. The test results of the product should not be used as the only basis for the individualized treatment of patients. Clinicians should comprehensively judge the test results in combination with the patient’s condition, drug indications, treatment response and other laboratory test indicators.
Anaplastic lymphoma kinase (ALK) wasfirst found as a subtype of anaplastic large cell lymphoma (ALCL). ALK gene islocated at chromosome 2p23. Normally, human ALK can be transcribed to produce 6222 BP mRNA, which is composed of 29 exons, encoding 1620 amino acid sequences and 200kda type I transmembrane protein. This protein is a receptor tyrosine kinase (RTK) and a member of RTK insulin superfamily. Subsequently, it was found that many types of ALK gene fusion were found in non-small cell lung cancer, diffuse large B-cell lymphoma and inflammatory myofibroblast tumor, which proved that ALK gene fusion was a strong carcinogenic driver gene.
At present, the incidence rate and mortality rate of lung cancer are first of the malignant tumors in China, of which 80%~85% is non-small- cell lung cancer (NSCLC). Individualized molecular targeted therapy based on epidermal growth factor receptor (EGFR) and anaplastic lymphoma kinase (ALK) has become a research hotspot of NSCLC. Many clinical studies have proved that ALK inhibitors can benefit NSCLC patients with ALK fusion gene positive. Therefore, it is of great clinical significance to detect ALK gene status in patients with NSCLC.
Based on the fluorescence in situ hybridization technology, a nucleotide of the nucleic acid probe is labeled with fluorescein. The detected target gene is homologous and complementary with the used nucleic acid probe. After denaturation, annealing and renaturation, they can form a hybrid between the target gene and the nucleic acid probe. The detected gene is analyzed qualitatively, quantitatively or relatively under the microscope by the fluorescence detection system. The kit adopts rhodamine fluorescein (rho) labeled orange probe and fluorescein isothiocyanate (FITC) labeled green probe. The two probes can be combined with the target detection site by in situ hybridization. Under normal conditions (ALK gene does not fuse), it is displayed as green and orange signals close to each other or yellow signals overlapping each other under fluorescence microscope. When there is gene fusion, the green and red signals are “broken” due to the replacement of recombinant fusion partner genes, and appear as signals far apart. The fusion of ALK gene in the tissues of patients with non-small cell lung cancer was detected by this broken fluorescence in situ hybridization method, so asto provide a reference basis for the treatment prognosis and medication of patients with non-small cell lung cancer.
This kit consists of ALK dual color probe, asshown in Table 1.
The reagents not provided in the kit are shown in Table 2.
| Component name | Specifications | Quantity | Main components |
|---|---|---|---|
| ALK dual color probe | 100μL/Tube | 1 | ALK Orange probe ; ALK Green probe |
| Reagent name | Purity | Reagent name | Purity |
|---|---|---|---|
| Sodium chloride | Analytical purity AR | NP-40 | Analytical purity AR |
| Sodium citrate | Analytical purity AR | Xylene | Analytical purity AR |
| Anhydrous ethanol | Analytical purity AR | Protease K | ≥40 units/g |
Keep sealed away from light at -20oC±5oC. The product is valid for 12 months. Avoid unnecessary repeated freezing and thawing that should not exceed 10 times. After opening, within 24 hours for short-term preservation, keep sealed at 2~8oC in dark. For long-term preservation after opening, keep the lid sealed at -20oC±5oC away from light. See the label of the kit for the production date and expiration date.
1. Related reagents
The following reagents are required for the experiment but not provided in this kit
Sodium chloride
176g
Sodium citrate
88g
Weigh 176g of sodium chloride and 88g of sodium citrate, dissolve in 800mL of deionized water, adjust the pH to 5.3±0.2 at room temperature, and complete to 1 L with deionized water. High-pressure steam sterilization, stored at 2~8oC, the solution shelf life is of 6 months. Discard if the reagent appears cloudy (turbid) or contaminated.
NP-40
0.6mL
20xSSC
4mL
Take 0.6mL NP-40 and 4mL 20×SSC, add 150mL deionized water, mix, adjust the pH to 7.0 ~ 7.5 atroom temperature, with deionized water complete to a volume of 200mL. Stored at 2~8oC, the shelf life is of 6 months. Discard if the reagent appears cloudy (turbid) or contaminated.
NP-40
0.2mL
20xSSC
20mL
Take 0.2mL NP-40 and 20mL 20×SSC, add 150mL deionized water, mix, adjust the pH 7.0±0.2 at room temperature, with deionized water complete to a volume of 200mL. Stored at 2~8oC, the shelf life is of 6 months. Discard if the reagent appears cloudy (turbid) or contaminated.
2. Pretreatment
3.Denaturation and Hybridization
The following operations should be performed in a darkroom.
4.Washing
The following operations should be performed in a darkroom.
5.Counterstaining
The following operations should be performed in a darkroom 10-15μl DAPI compound dye is dropped in the hybridization area of the glass slide and immediately covered. The suitable filter is selected for glass slide observation under the fluorescence microscope.
6.FISH results observation
Place the slides under the fluorescence microscope after counterstaining, then under the natural light, use a low-power objective (10x) to locate the NSCLC cell area, and then switch to 40× objective to locate an area where cells are well-distributed. Use high-power objective (60x, 100x) to select the cells which have same size of nuclei, complete nuclei boundary, well DAPI staining, no overlapping of nuclei and show clear signal, randomly choose at least 50 tumor cells, enumerate the orange and green signal in these nuclei.

Individual orange or green signals are considered assingle signals.
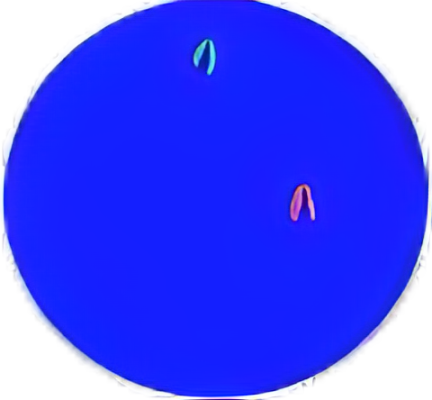
Diffuse signals can have a fuzzy or elongated DNA fiber appearance.
Orange and green signals are adjacent that the distance is less than two signal diameters, or are overlapping, which are considered as one fused signal. Multiple fused and/ or broken apart signals may be observed in a single nucleus.

If diffuse signal is adjacent or connected with a fiber signal, they should be recorded as one fused signal. Multiple fused and/or broken apart signals may be observed in a single nucleus.

Two signals of the same color that have the same size are adjacent, separated by a distance less than two signal diameters, which should be recorded as one signal, (this is a split signal).
Enumerate 50 tumor cells, record the number of fused or adjacent signals, single orange signal and single green signal in each cell. Enumerate once for each tumor cell, and record the cells with hybridization signal (both orange and green signal), but these cells with no signal, only single color signal, weak signal or extremely diffuse signal should not be counted.
Results determination
Negative signal (No gene fusion) |
|
|---|---|
|
|
A. and B. These nuclei contain fused orange and green signals. The signals are either overlapping, adjacent, or are less than two signal diameters apart. C. A single green signal without a corresponding orange signal in addition to fused and/or broken-apart signals indicates a deletion of the orange portion of the ALK probe and is considered negative. The target area of the drug is located within the area targeted by the orange probe. |
Positive signal (Gene fusion) |
|
|---|---|
|
|
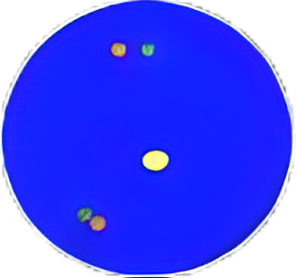
Positive signal: these nuclei contain rearranged or “broken apart” signals, 2 or more signal diameters apart. Figure a. A nucleus can have more than one set of broken apart signals. Figure b. A nucleus can have fused signal (s) and broken apart signal (s). Figure c. A nucleus can have a single orange signal (deleted green signal) in addition to fused and/or broken apart signals. Note: A nucleus with signals of only one color should not be enumerated. Figure d. The same nucleus has fused signals, broken apart signals and deletions. |
| Signal type | Number of adjacent or fused signals | Number of single orange signals | Number of single green signals | Cell determination |
|---|---|---|---|---|
|
A、B |
≥1 |
0 |
0 |
Negative |
|
C |
≥1 |
0 |
≥1 |
Negative |
|
a、b、d |
≥0 |
≥1 |
≥1 |
Positive |
|
c |
≥1 |
≥1 |
0 |
Positive |
Enumerate the positive cells and negative cells based on the determination methods described in Table 4 and 5, calculate Ratio value, Ratio = number of positive cells/ total number of enumerated cells × 100%.
| Problem | Probable cause | Recommended solution |
|---|---|---|
| Strong background of slides | Inadequate wash of glass slide before preparation of specimens | Wash the glass slide using the absolute ethyl alcohol. |
| Inadequate wash after hybridization | Assure that the wash buffer is prepared in line with Instruction For Use, assure the correct pH value and temperature of wash buffer, remove the coverslip and repeat the washing steps. |
|
| Improper use of filter sets | Replace with suitable filter sets to reduce the background light. | |
| Improper hybridization condition | Assure the temperature of hybridization instrument is set as 42ºC. | |
| The temperature is too low when washing / The washing intensity of wash buffer is too low | Assure that the wash buffer reaches the required temperature when washing the slides. Assure the wash buffer is prepared in line with Instruction For Use. (low SSC concentration or high NP-40 concentration would help improving the washing intensity of wash buffer). | |
| The washing intensity of wash buffer is too low | Assure the wash buffer is prepared in line with Instruction For Use. (low SSC concentration or high NP-40 concentration would help improving the washing intensity of wash buffer). | |
| Weak counterstaining | Weak counterstaining | Remove coverslip, at room temperature, immerse the slides in the wash buffer containing 2 × SSC/0.1%NP-40 for 5 minutes. Then sequentially immerse the slides in 70%, 85% and 100% ethanol solution for 1 minute respectively, and then perform the counterstaining. |
| The counter stain has been kept under long-term storage or excessive light | Assure the counter stain is stored at -20ºC and protected away from light, assure its effect. | |
| No signal or weak signals | Inadequate denaturation of specimens | Assure the temperature of hybridization instrument is set as 83ºC, at least 10 minutes in advance is needed to preheat hybridization instrument. |
| The probe mixture and hybridization buffer were not mixed sufficiently before use | Blow the probe mixture and mix the probe sufficiently, centrifuge for a short time. | |
| The probe mixture on tissue slides dries too fast | After dropping probe mixture the target area should be covered by coverslip immediately, when washing the slides you can only remove one coverslip at a time, and dip it into wash buffer immediately before removing next coverslip. | |
| Air bubbles formed under coverslip during hybridization | The coverslip should cover the probe mixture in order to gently squeeze out air bubbles. | |
| Inappropriate hybridization condition | Ensure to comply with the time and temperature required by hybridization and do not leave gaps when sealing the slides with rubber cement. The hybridization time should be adjusted according to the situation. | |
| Improper wash buffer or incorrect washing conditions | Be sure to follow the requirements of Instruction for Use to formulate the wash buffer. Ensure that the temperature of wash buffer reaches the temperature predetermined in washing step. The thermometer and pH meter should be accurately calibrated. Remove coverslip before immersing the slide into wash buffer. | |
| Inappropriate storage of probe or specimens slides | Make sure that the probe mixture is stored at -20ºC and protected from light. Place the slides without hybridization at -20ºC for long-term storage or at room temperature for short-term storage. Place the hybridized slides at -20ºC, away from light, and store for less than 6 months. | |
| Incorrect use of DAPI counter stain, excessively high brightness of counter stain | Remove the coverslip, immerse the slides in 2 × SSC/ 0.1%NP-40 for 5 minutes at room temperature. Sequentially immerse the slides in 70%, 85% and 100% ethanol solution for 1 minute respectively, and then perform the counterstaining after air drying the slides. | |
| Inappropriate filter sets were selected for observation | Use correct filter sets to observe the probe fluorescence. For the detailed information, please consult the technical service department of Gene Bio Solution. |
The clinical study of this kit adopts the experimental design of a synchronous blind method and uses the concomitant diagnostic reagent (Abbott Molecular’s “ALK gene recombination detection kit (fluorescence in situ hybridization)” (Registration Certificate No.: GXZZ 20143405183) that has been verified by targeted drugs as the comparison reagent. A total of 1189 effective samples have been tested, and the positive coincidence rate of this kit, the negative coincidence rate, and the overall coincidence rate were 100%, and the kappa value was 1.000 (P = 0.00).
The results of this kit will be affected by various factors of the sample itself, but also limited by enzyme digestion time, hybridization
temperature and time, operating environment and the limitations of current molecular biology technology, which may lead to wrong ALK gene fusion results. Users must understand the potential errors and accuracy limitations that may exist in the detection process.
