Product Name
TERC gene amplification probe detection kit.
Package Specifications
10 Tests/box
Intended Use
This kit uses fluorescence in situ hybridization to detect TERC gene status, and the test sample is cervical exfoliated cells.
Cervical cancer is one of the most common malignant tumors in the female reproductive system. The incidence rate of cervical cancer is the second place in the world, which seriously threatens the health of women. Understanding the molecular basis of tumorigenesis and determining genetic markers by studying the genetic changes of cervical cancer is of great significance to early predict the prognosis of high-risk patients. Studies have shown that the occurrence and development of cervical cancer is often accompanied by chromosome 3q amplification, located at 3q26 Human telomerase gene (hTERC) in region 3 can be amplified by chromosome 3q amplification. Studies have confirmed that the abnormal change of TERC is the necessary condition for the development of cervical intraepithelial neoplasia to cervical cancer, and the positive rate of TERC gene will increase with the increase of cervical lesion level. Therefore, it has a certain predictive significance for the progression of cervical intraepithelial neoplasia.
TERC gene detection can detect TERC gene amplification in cervical precancerous lesions, so TERC gene detection can be used for early diagnosis of cervical cancer, and TERC gene amplification level is positively correlated with tumor malignancy. It is feasible to assist in the diagnosis of high-grade cervical intraepithelial lesions, and also has important clinicalsignificance for the identification ofsurgical treatment indications, It can predict the development trend of cervical precancerous lesions to a certain extent, and can be a good supplement to the examination of cervical cell morphology and molecular cytogenetics, so as to provide a new way for the screening and prognosis evaluation of cervical cancer and precancerous lesions.
This kit only verifies the detection performance of TERC gene. This kit is only applicable to the detection of TERC gene status and provides doctors with auxiliary information for diagnosis.
Detection principle
Fluorescence in situ hybridization is a technique for directly observing specific nucleic acids in cells. According to the principle of base complementary pairing, the specific DNA sequence is complementary to the target sequence in the cell. Due to the fluorescence of the probe, the hybrid probe and target DNA can be clearly observed under the fluorescence microscope under the appropriate excitation light. Hybridization includes the following steps: first, preprocess the samples according to the instructions; Secondly, DNA was denatured and hybridized with the probe; After hybridization, the excess unbound probes were washed away through a series of washing, and the nuclei were counterstained blue with DAPI (4,6-diamidino-2-phenylindole hydrochloric acid); Finally, the fluorescence signals from DAPI and the probe were observed under a suitable filter by fluorescence microscope.
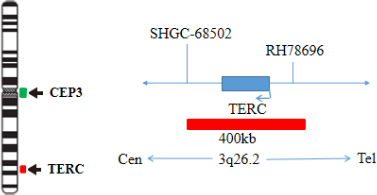
The kit uses orange fluorescein labeled TERC probe to detect TERC gene, and green fluorescein labeled cep3 probe to detect the centromere sequence of chromosome 3. The two probes can be combined with the target detection site by in situ hybridization. The signal number corresponding to cep3 probe reflects the number of chromosome 3 at the target site, and the signal number of TERC probe corresponds to the status of TERC gene at the target site. The amplification status of TERC gene in the cells to be detected can be determined by the ratio of orange signal to green signal fluorescence number.
Product Composition
The kit consists of TERC/ CEP3 dual-color probes, as shown in Table 1.
Storage conditions & Validity
Keep sealed away from light at -20oC±5oC. The product is valid for 20 months. Avoid unnecessary repeated freezing and thawing thatshould not exceed 10 times. After opening, within 24 hours for short-term preservation, keep sealed at 2-8oC in dark. For long-term preservation after opening, keep the lid sealed at -20oC±5oC away from light. The kit is transported under 0°C.
Applicable Instruments
- Fluorescence microscopy imaging system includes fluorescence microscopy and filter setsfor DAPI, Green, and Orange.
Sample Requirements
- Applicable specimen types: Cervical exfoliated cells (TCT production).
- Samples should be disposed of promptly (usually not overnight) and should be discarded if the storage temperature is too high or too
low (eg, frozen).
Test method
-
- Related reagents
The following reagents are required for the experiment but not provided in this kit
- 20×SSC, pH 5.3±0.2
Weigh 176g of sodium chloride and 88g of sodium citrate, dissolve in 800mL of deionized water, adjust the pH to 5.3±0.2 at room temperature, and complete to 1 L with deionized water. High-pressure steam sterilization, stored at 2-8°C, the solution shelf life is of 6 months. Discard if the reagent appears cloudy (turbid) or contaminated.
- 2×SSC, pH 7.0±0.2
Take 100mL of the above 20xSSC, dilute with 800mL deionized water, mix, adjust the pH to 7.0±0.2 at room temperature, complete to 1L with deionized water, stored at 2-8°C, the shelf life is of 6 months. Discard if the reagent appears cloudy (turbid) or contaminated.
- Ethanol Solution: 70% ethanol, 85% ethanol
Dilute 700ml, 850ml of ethanol with deionized water to 1L. The shelf life is of 6 months. Discard if the reagent appears cloudy (turbid) or
contaminated.
- 0.3% NP-40/0.4xSSC solution, pH 7.0-7.5
Take 0.6mL NP-40 and 4mL 20×SSC, add 150mL deionized water, mix, adjust the pH to 7.0-7.5 at room temperature, with deionized water complete to a volume of 200mL. Stored at 2-8°C, the shelf life is of 6 months. Discard if the reagent appears cloudy (turbid) or contaminated.
- 0.04% pepsin
Preparation of 2% pepsin solution: Weigh accurately and dissolve in 10 ml of deionized water 0.20g of pepsin (sigma).
Preparation of 0.04% pepsin solution: Measure 50ml of deionized water, add 95μl of concentrated hydrochloric acid, mix well, add 1ml of the prepared 2% pepsin solution, and mix gently. Preheat at 37°C before each use.
- 0.1M HCl solution
Measure 8.2ml concentrated HCL, mix with deionized water and complete to 100ml, store at room temperature to obtain 1M HCL. Based on needs dilute to 0.01M by 10-fold dilution method.
- Di-amiindyl phenyl indole (DAPI) dyeing agent
Please use commercially available DAPI counterstains containing anti-quencher.
- Slides pretreatment process
- Acidification: Rinse twice slides with 2xSSC at room temperature for 5 min each time and rinse once with 0.1M HCL for 10 min.
- Digestion: Digest with 0.04% pepsin at 37°C for 10 min, and rinse twice the slides win 2xSSC solution at room temperature for 5 min each time.
- Dehydration: Place slides in -20°C pre-cooled 70% ethanol, 85% ethanol and 100% ethanol for 2 min each for dehydration. Dry slides naturally.
- Carry out hybridization experiment according to the hybridization procedure.
- Fixation: Aspirate the supernatant, add 5 mL of freshly prepared 3:1 methanol and glacial acetic acid fixative solution, mix with a pipette, fix for 10 min, and centrifuge at 1000 rpm for 10 min.
- Denaturation and Hybridization
The following operations should be performed in a darkroom.
- Take the probe at room temperature for 5 minutes. Briefly centrifuge manually (do not use vortex or shaker instrument). Take 10μL droplet in the cell and drop in the hybridization zone, immediately cover 22mmx22mm glass slide area; spread evenly without bubbles the probe under the glass slide covered area and seal edges with rubber (edge sealing must be thorough to prevent dry film from affecting the test results during hybridization).
- Place the glass slide in the hybridization instrument, denature at 88°C for 2 minutes (the hybridizer should be preheated to 88°C) and hybridize at 45°C for 2 to 16 hours.
- Washing
The following operations should be performed in a darkroom.
- Take out the hybridized glass slides, remove the rubber on the coverslip and immediately place the slides into 2xSSC for 5 seconds, and gently remove the coverslip.
- Place the glass slides in 2xSSC at room temperature for 1 min.
- Remove and immerse the slides in a 0.3% NP-40/0.4×SSC solution preheated at 68°C for 2 min. (Preparation of 0.3% NP-40/0.4xSSC: For 1L preparation, take 3mL NP-40 and 20mL 20xSSC, dissolve fully, mix well, and use 1M NaOH to adjust the pH to 7.2).
- Immerse the glass slides in deionized water at 37°C for 1min, and dry naturally in the dark.
- Counterstaining
The following operations should be performed in a darkroom.
10-15μl DAPI compound dye is dropped in the hybridization area of the glass slide and immediately covered for 10-20min. The suitable filter is selected for glass slide observation under the fluorescence microscope.
- FISH results observation
Place the stained sections under a fluorescence microscope and the cells area is first confirmed under a low magnification objective (10x); under magnification objective (40x) a uniform cells distribution is observed; then the nucleus size uniformity, nuclear boundary integrity, DAPI staining uniformity, no nuclei overlapping, cells clear signal are observed in the high magnification objective (60x, 100x). Select randomly 100 tumor cells at least and count the orange and green signals in the nuclei.
Positive judgment value or reference interval
- Signal classification and counting
- Normal cell signal: there are 2 orange red signals and 2 green signals in a single interphase nucleus.
- Abnormal cell signals: the number of orange signals in a single interphase nucleus > 2 and the number of green signals ≥ 2.
100 cells were randomly counted, and the number of normal signal cells and abnormal signal cells were counted respectively. Each cell is counted once. Only cells with hybridization signal (both color signals) are counted. Cells without signal or only a single color signal are not counted. Cells with weak signal or too diffuse signal are not counted.
- FISH result judgment
To judge the abnormality of the detection result, it is necessary to establish the abnormality threshold
- Abnormal threshold establishment
- It is recommended to select 20 normal cervical samples as negative control.
- The slides were prepared by the above methods and steps for fish experiment.
- Establishment of abnormal threshold: analyze 100 cells per sample and calculate the average value and standard deviation of the
percentage of cells showing abnormal signal mode. The abnormal threshold is defined as the average value + 3 × Standard deviation.
Anomaly threshold = mean (m) + 3 × Standard deviation (SD)
Example: Table 2: select 20 normal cervical samples as negative control for fish detection.
Table 2: Abnormal threshold setup
| No. |
Abnormal cells (%) |
|
Sample 1
Sample 2
……………
Sample 20
Mean
SD
Threshold Value
|
5
3
.........
4
3
0.3
(Anomaly threshold = Mean value + 3 x SD) = 3.9
|
2. Result judgment:
If the detection value of the number of cells displaying abnormal signal mode is greater than the abnormal threshold, it is determined as a positive result; If the detection value of the number of cells displaying abnormal signal mode is less than the abnormal threshold, it is determined as a negative result; If the detection value of the number of cellsin the abnormalsignal mode is equal to the abnormal threshold, increase the number of observation sample cells and count 200 cells to judge the final result.
Taking Table 2 as an example, if the percentage of abnormal signal cells in the fish test results of the sample to be tested is greater than 3.9%, that is, the abnormal threshold (e.g. 8%), it isjudged that there is TERC gene amplification in the sample; If the percentage of abnormal signal cells in the fish test results of the sample to be tested is less than 3.9%, that is, the abnormal threshold (e.g. 2%), it is judged that there is no TERC gene in the sample.
Product performance index
- Fluorescence signal intensity: after the probe is effectively hybridized with the karyotype sample, itshould emit a fluorescence signal that can be recognized by the naked eye under the fluorescence microscope.
- Sensitivity: detect karyotype samples, analyze the chromosomes of 50 cellsin metaphase, and at least 98 chromosomes 17 show 1 orange fluorescence signal and 1 green fluorescence signal.
- Specificity: Analyze the chromosomes of 50 cells in metaphase, at least 98 chromosomes 17 show a specific orange fluorescence signal in the target region and a specific green fluorescence signal in the centromere region.
Precautions
- Please read this manual carefully before testing. The testing personnel should receive professional technical training. The signal counting personnel must be able to observe and distinguish orange and green signals.
- When testing clinical samples, when it is difficult to count the hybridization signals and the samples are not enough to repeat the test, the test will not provide any test results. If the number of cells is not enough for analysis, the test will not provide test results.
- DAPI counterstaining agent used in this experiment has potential toxicity or carcinogenicity, so it is necessary to operate in the fume hood, wear masks and gloves to avoid direct contact.
- The results of this kit will be affected by various factors of the sample itself, but also limited by enzyme digestion time, hybridization temperature and time, operating environment and the limitations of current molecular biology technology, which may lead to wrong interpretation results. Users must understand the potential errors and accuracy limitations that may exist in the detection process.
- All chemicals are potentially dangerous. Avoid direct contact. Used kits are clinical waste and should be properly disposed off.

